OMERO.FPBioimage
In this document, we introduce OMERO.FPBioimage, a 3D volume viewer for OMERO.web.
Description:
We will show here:
How to open a multi-z image in OMERO.FPBioimage
Resources:
Example files used
Note: Only some of the images in this dataset are z-stacks, for example
Setup:
OMERO.FPBioimage installation
OMERO.FPBioimage is a pip installable application for OMERO.web. Follow the steps described in https://pypi.org/project/omero-fpbioimage/ to install it and configure the OMERO.web accordingly.
Step-by-Step:
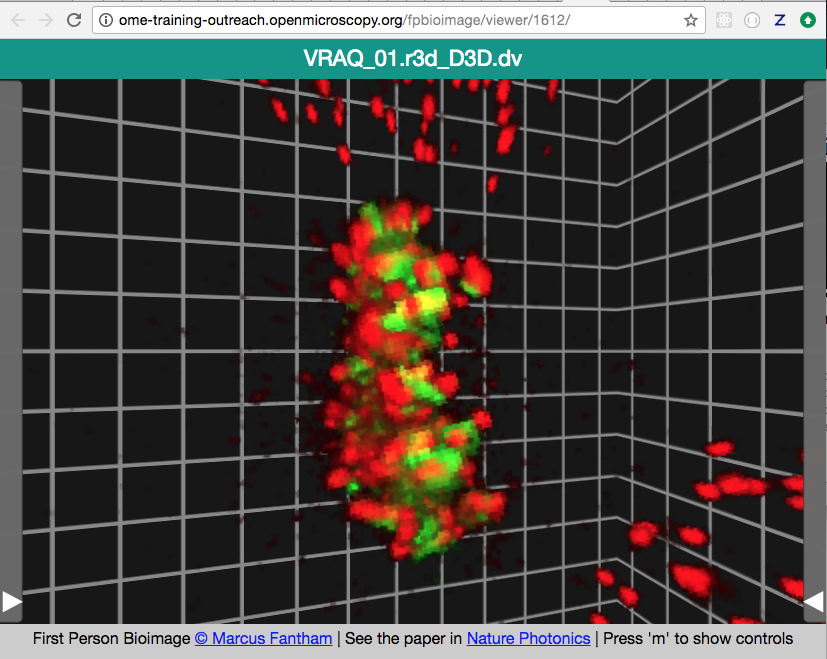
Login to OMERO.web and open an image from the Dataset siRNA-HeLa with multiple Z-sections e.g. VRAQ_01.r3d_D3D.dv in a 3D viewer: OMERO.FPBioimage.
First select the Image.
In the Preview tab, switch off all channels except FITC and the GFP-INCENP channel.
Save the new rendering settings.
Use right-click menu on the image in the left panel, or the Open with… icon
 on top of the right-hand pane to open the image with FPBioimage.
on top of the right-hand pane to open the image with FPBioimage.Click Start in the new viewer window.
We can see that the centromeres are well aligned on the metaphase plate on the selected Image, whereas the centromeres are located in and around the spheroid on the IN_02.r3d Image for example.